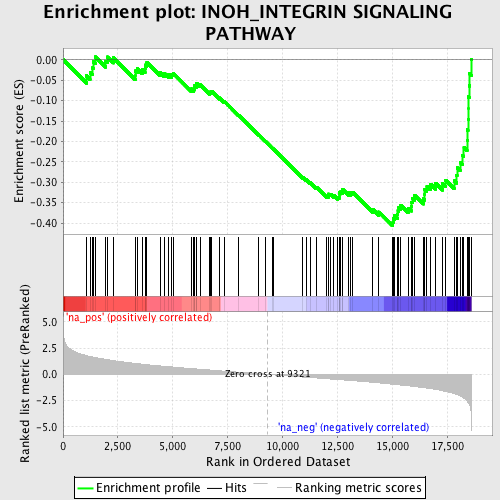
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | INOH_INTEGRIN SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.4070686 |
| Normalized Enrichment Score (NES) | -1.8262259 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.684838 |
| FWER p-Value | 0.664 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ITGB2 | 1063 | 1.780 | -0.0391 | No | ||
| 2 | SOS1 | 1228 | 1.701 | -0.0304 | No | ||
| 3 | SOS2 | 1347 | 1.650 | -0.0199 | No | ||
| 4 | HRAS | 1373 | 1.641 | -0.0043 | No | ||
| 5 | CDC42 | 1468 | 1.605 | 0.0071 | No | ||
| 6 | NCAN | 1936 | 1.421 | -0.0035 | No | ||
| 7 | DOCK3 | 2010 | 1.393 | 0.0069 | No | ||
| 8 | RHOA | 2286 | 1.305 | 0.0054 | No | ||
| 9 | GPC4 | 3286 | 1.040 | -0.0378 | No | ||
| 10 | ITGAL | 3291 | 1.039 | -0.0273 | No | ||
| 11 | MAPK3 | 3384 | 1.017 | -0.0218 | No | ||
| 12 | ITGB7 | 3605 | 0.966 | -0.0238 | No | ||
| 13 | VAV1 | 3736 | 0.936 | -0.0212 | No | ||
| 14 | TLN1 | 3759 | 0.932 | -0.0128 | No | ||
| 15 | LAMB3 | 3818 | 0.920 | -0.0065 | No | ||
| 16 | ITGAE | 4434 | 0.800 | -0.0314 | No | ||
| 17 | DCN | 4631 | 0.760 | -0.0342 | No | ||
| 18 | ITGA4 | 4815 | 0.729 | -0.0366 | No | ||
| 19 | ITGAM | 4942 | 0.706 | -0.0361 | No | ||
| 20 | SHC1 | 5008 | 0.694 | -0.0325 | No | ||
| 21 | LCK | 5834 | 0.552 | -0.0713 | No | ||
| 22 | KERA | 5951 | 0.530 | -0.0722 | No | ||
| 23 | GSG2 | 5988 | 0.524 | -0.0687 | No | ||
| 24 | COL11A2 | 5994 | 0.524 | -0.0636 | No | ||
| 25 | COL11A1 | 6069 | 0.509 | -0.0624 | No | ||
| 26 | COL12A1 | 6079 | 0.506 | -0.0576 | No | ||
| 27 | COL6A2 | 6241 | 0.483 | -0.0614 | No | ||
| 28 | ITGA3 | 6683 | 0.410 | -0.0810 | No | ||
| 29 | SRC | 6721 | 0.404 | -0.0788 | No | ||
| 30 | ITGB3 | 6756 | 0.399 | -0.0765 | No | ||
| 31 | COL6A1 | 7132 | 0.337 | -0.0933 | No | ||
| 32 | LUM | 7359 | 0.299 | -0.1024 | No | ||
| 33 | SDC2 | 8011 | 0.203 | -0.1355 | No | ||
| 34 | ITGA2B | 8920 | 0.059 | -0.1839 | No | ||
| 35 | DOCK4 | 9222 | 0.016 | -0.2000 | No | ||
| 36 | CAV1 | 9549 | -0.034 | -0.2172 | No | ||
| 37 | MAP2K1 | 9587 | -0.041 | -0.2188 | No | ||
| 38 | DOCK2 | 10910 | -0.254 | -0.2876 | No | ||
| 39 | BCAR1 | 11071 | -0.278 | -0.2933 | No | ||
| 40 | BGN | 11261 | -0.306 | -0.3004 | No | ||
| 41 | COL10A1 | 11546 | -0.343 | -0.3122 | No | ||
| 42 | COL1A1 | 12026 | -0.413 | -0.3338 | No | ||
| 43 | ELN | 12081 | -0.422 | -0.3324 | No | ||
| 44 | COL4A2 | 12096 | -0.426 | -0.3288 | No | ||
| 45 | ITGA6 | 12181 | -0.438 | -0.3288 | No | ||
| 46 | ITGAX | 12338 | -0.457 | -0.3325 | No | ||
| 47 | COL8A1 | 12506 | -0.484 | -0.3366 | No | ||
| 48 | DOCK9 | 12581 | -0.495 | -0.3355 | No | ||
| 49 | ITGB6 | 12585 | -0.496 | -0.3305 | No | ||
| 50 | RAC1 | 12586 | -0.496 | -0.3254 | No | ||
| 51 | ITGA11 | 12650 | -0.506 | -0.3236 | No | ||
| 52 | SDC1 | 12730 | -0.519 | -0.3225 | No | ||
| 53 | DOCK7 | 12740 | -0.520 | -0.3177 | No | ||
| 54 | ITGA5 | 13004 | -0.565 | -0.3261 | No | ||
| 55 | MAPK9 | 13114 | -0.579 | -0.3260 | No | ||
| 56 | COL2A1 | 13202 | -0.594 | -0.3246 | No | ||
| 57 | KRAS | 14116 | -0.757 | -0.3661 | No | ||
| 58 | CRK | 14395 | -0.808 | -0.3728 | No | ||
| 59 | GRB2 | 15031 | -0.939 | -0.3974 | Yes | ||
| 60 | SMC3 | 15044 | -0.942 | -0.3884 | Yes | ||
| 61 | NRAS | 15105 | -0.954 | -0.3818 | Yes | ||
| 62 | COL8A2 | 15237 | -0.976 | -0.3788 | Yes | ||
| 63 | COL4A1 | 15262 | -0.981 | -0.3701 | Yes | ||
| 64 | SHC3 | 15300 | -0.993 | -0.3618 | Yes | ||
| 65 | MAP2K2 | 15389 | -1.012 | -0.3562 | Yes | ||
| 66 | RAP1A | 15728 | -1.081 | -0.3633 | Yes | ||
| 67 | VCAN | 15863 | -1.117 | -0.3591 | Yes | ||
| 68 | COL1A2 | 15883 | -1.124 | -0.3485 | Yes | ||
| 69 | RAP1B | 15926 | -1.135 | -0.3391 | Yes | ||
| 70 | COL9A1 | 16017 | -1.158 | -0.3321 | Yes | ||
| 71 | ITGAV | 16412 | -1.257 | -0.3404 | Yes | ||
| 72 | ACAN | 16476 | -1.274 | -0.3307 | Yes | ||
| 73 | VCL | 16483 | -1.276 | -0.3179 | Yes | ||
| 74 | BCAN | 16583 | -1.305 | -0.3099 | Yes | ||
| 75 | SDC3 | 16748 | -1.357 | -0.3048 | Yes | ||
| 76 | COL3A1 | 16974 | -1.426 | -0.3023 | Yes | ||
| 77 | FN1 | 17290 | -1.556 | -0.3033 | Yes | ||
| 78 | DOCK8 | 17439 | -1.620 | -0.2946 | Yes | ||
| 79 | COL18A1 | 17822 | -1.821 | -0.2965 | Yes | ||
| 80 | LAMC2 | 17930 | -1.892 | -0.2828 | Yes | ||
| 81 | MAPK10 | 17957 | -1.913 | -0.2646 | Yes | ||
| 82 | DOCK10 | 18096 | -2.048 | -0.2510 | Yes | ||
| 83 | FYN | 18200 | -2.163 | -0.2343 | Yes | ||
| 84 | HSPG2 | 18270 | -2.263 | -0.2148 | Yes | ||
| 85 | ITGA7 | 18412 | -2.509 | -0.1966 | Yes | ||
| 86 | ITGB5 | 18422 | -2.526 | -0.1711 | Yes | ||
| 87 | MAPK8 | 18456 | -2.662 | -0.1455 | Yes | ||
| 88 | ITGB1 | 18465 | -2.685 | -0.1183 | Yes | ||
| 89 | CD44 | 18469 | -2.701 | -0.0907 | Yes | ||
| 90 | RAF1 | 18508 | -2.852 | -0.0635 | Yes | ||
| 91 | SDC4 | 18530 | -2.957 | -0.0342 | Yes | ||
| 92 | PTK2 | 18604 | -3.772 | 0.0006 | Yes |
